PanPA
PanPA is a command line tool written in Cython for building and alignments of panproteome graphs. The code base can be found Here.
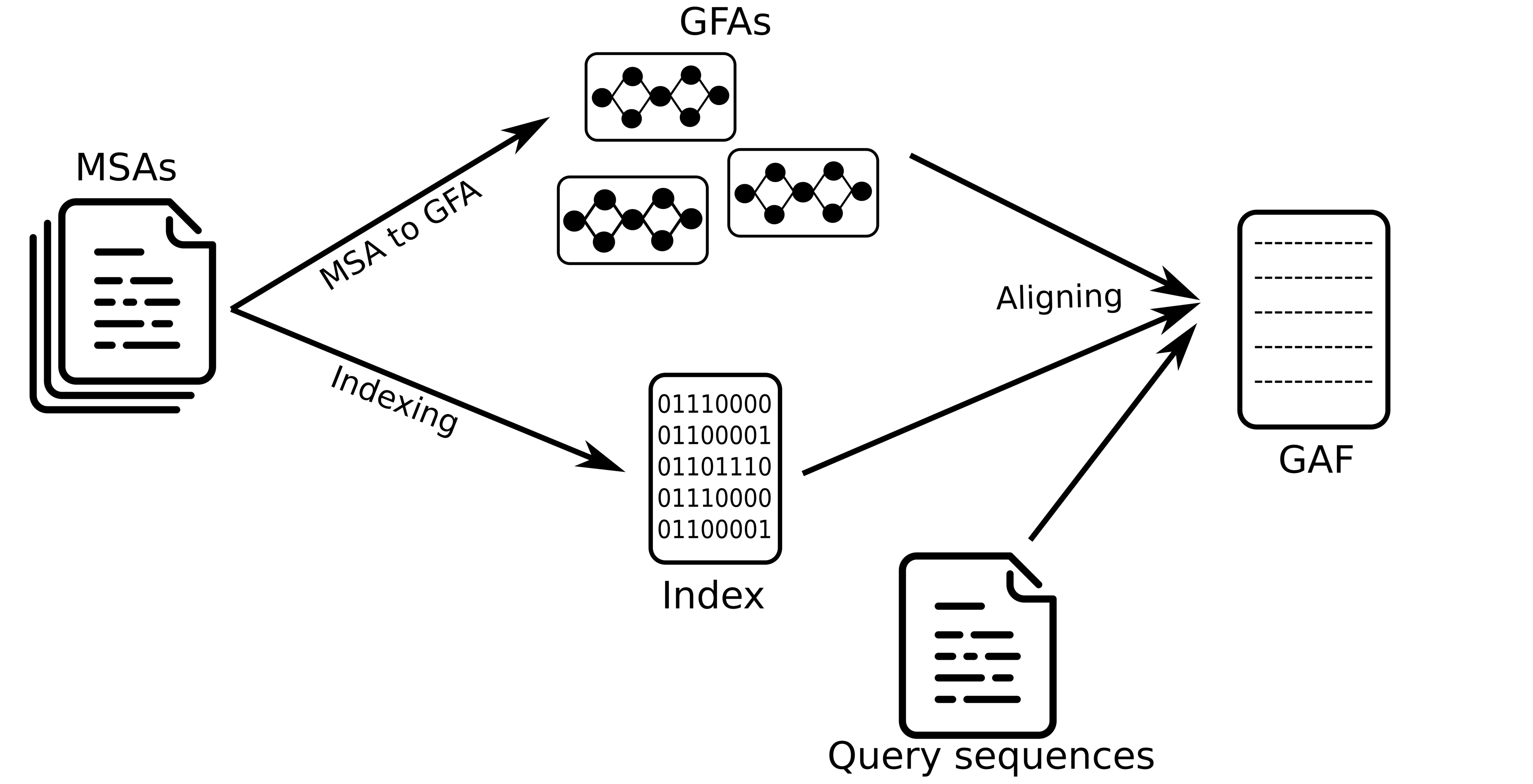
The idea here is that given a set of MSAs of protein sequences (whether it is the same protein or a protein cluster), each MSA is turned into a Directed Acyclic Graph (DAG) in GFA format, indexes each MSA using k-mers or (w, k)-minimizers, and align DNA and amino acid sequences back to these graph using the index to find matches to the graph.
The alignment is done using Partial Order Alignment algorithm and the user can choose different substitution matrices and gap penalty score.
More on usage and commands can be found in Subcommands
Installation
PanPA is easy to install through the setup.py script, the only requirement is Cython and Python >= 3.6. You can install PanPA locally with python3 setup.py install --user if you do not have root access to the operating system you’re working on.
You can also use the environment.yml file to generate a conda or virtual Python environment and install PanPA there.